|
||||||||
Bayesian Networks - Python Automation and Machine Learning for ICs - - An Online Book - |
||||||||
| Python Automation and Machine Learning for ICs http://www.globalsino.com/ICs/ | ||||||||
| Chapter/Index: Introduction | A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Appendix | ||||||||
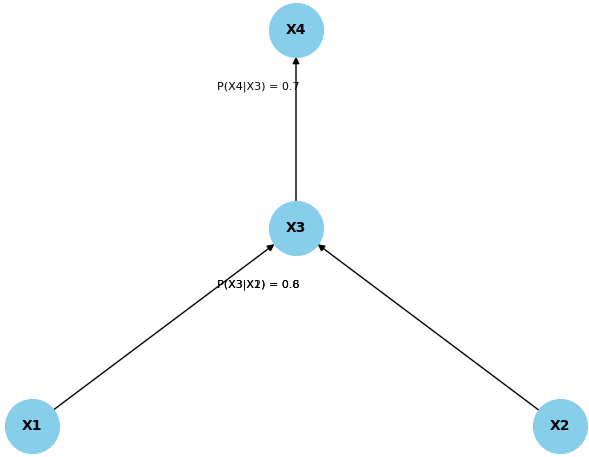
================================================================================= In machine learning, particularly in probabilistic graphical models or Bayesian statistics, conditional probabilities play a significant role. A Bayesian network is indeed a data structure that represents the dependencies among random variables. It is a probabilistic graphical model that uses a directed acyclic graph (DAG) to depict the relationships between variables. Nodes in the graph represent random variables, and directed edges between nodes indicate probabilistic dependencies. That is, in Bayesian networks, nodes represent random variables, and the edges between them indicate probabilistic dependencies. In addition to the graph structure, Bayesian networks also include conditional probability tables (CPTs) associated with each node, which describe the probabilistic relationship between a node and its parents in the graph. This allows Bayesian networks to model complex probabilistic dependencies in a compact and interpretable way. The conditional probabilities P(xi|parents(xi)) specify how each variable depends on its parents in the graph. In DAG, the structure of a Bayesian network is a directed graph where nodes represent random variables, and directed edges (arrows) between nodes represent probabilistic dependencies. The graph is acyclic, meaning there are no cycles or loops. Each node in the graph corresponds to a random variable. These random variables can represent observable data or latent variables (unobserved factors).The directed edges in the graph indicate probabilistic dependencies between the connected nodes. If there is an arrow from node X to node Y, it means that X is considered a parent of Y, implying that Y's probability distribution depends on the values of X. Associated with each node is a conditional probability table (CPT). The CPT specifies the probability distribution of a node given its parents in the graph. It quantifies how the variable's values depend on the values of its parent variables. Figure 3818 shows a Bayesian network with conditional probabilities. The values for conditional probabilities reflect the strength or likelihood of the dependency between the variables in the Bayesian network. These probabilities are often estimated from training data or domain knowledge. In practice, the actual value of the conditional probabilities would depend on the specific context and the information available during the model training phase.
============================================
|
||||||||
| ================================================================================= | ||||||||
|
|
||||||||